---
title: "Lab 1.4: Data Representations"
output:
BiocStyle::html_document:
toc: true
vignette: >
% \VignetteIndexEntry{Lab 1.5: Data Represenations}
% \VignetteEngine{knitr::rmarkdown}
---
```{r style, echo = FALSE, results = 'asis'}
BiocStyle::markdown()
```
```{r setup, echo=FALSE}
knitr::opts_chunk$set(
eval=as.logical(Sys.getenv("KNITR_EVAL", "TRUE")),
cache=as.logical(Sys.getenv("KNITR_CACHE", "TRUE"))
)
suppressPackageStartupMessages({
library(Biostrings)
library(GenomicRanges)
})
```
Original Authors: Martin Morgan, Sonali Arora
Presenting Authors: [Martin Morgan][], [Lori Shepherd][]
Date: 22 July, 2019
Back: [Monday labs](lab-1-intro-to-r-bioc.html)
[Martin Morgan]: mailto: Martin.Morgan@RoswellPark.org
[Lori Shepherd]: mailto: Lori.Shepherd@RoswellPark.org
**Objective**: Learn the essentials of _Bioconductor_ data structures
**Lessons learned**:
- Review of common bioinformatic file formats: FASTA, FASTQ, SAM /
BAM, VCF, BED / WIG / GTF
- How to work with _Bioconductor_ objects for DNA sequences, genomic
ranges, aligned reads, and called variants.
# Classes, methods, and packages
This section focuses on classes, methods, and packages, with the goal
being to learn to navigate the help system and interactive discovery
facilities.
## Motivation
Sequence analysis is specialized
- Large data needs to be processed in a memory- and time-efficient manner
- Specific algorithms have been developed for the unique
characteristics of sequence data
Additional considerations
- Re-use of existing, tested code is easier to do and less error-prone
than re-inventing the wheel.
- Interoperability between packages is easier when the packages share
similar data structures.
Solution: use well-defined _classes_ to represent complex data;
_methods_ operate on the classes to perform useful functions. Classes
and methods are placed together and distributed as _packages_ so that
we can all benefit from the hard work and tested code of others.
## Objects
Load the [Biostrings][] and [GenomicRanges][] package
```{r setup-objects}
library(Biostrings)
library(GenomicRanges)
```
- _Bioconductor_ makes extensive use of classes to represent
complicated data types
- Classes foster interoperability -- many different packages can work
on the same data -- but can be a bit intimidating for the user.
- Formal 'S4' object system
- Often a class is described on a particular home page, e.g.,
`?GRanges`, and in vignettes, e.g.,
`vignette(package="GenomicRanges")`,
`vignette("GenomicRangesIntroduction")`
- Many methods and classes can be discovered interactively , e.g.,
`methods(class="GRanges")` to find out what one can do with a
`GRanges` instance, and `methods(findOverlaps)` for classes that
the `findOverlaps()` function operates on.
- In more advanced cases, one can look at the actual definition of
a class or method using `getClass()`, `getMethod()`
- Interactive help
- `?findOverlaps,` to select help on a specific method,
`?GRanges-class` for help on a class.
## Example & short exercise: _Biostrings_
Example: _Biostrings_ for DNA sequences
```{r Biostrings, message=FALSE}
library(Biostrings) # Biological sequences
data(phiX174Phage) # sample data, see ?phiX174Phage
phiX174Phage
m <- consensusMatrix(phiX174Phage)[1:4,] # nucl. x position counts
polymorphic <- which(colSums(m != 0) > 1)
m[, polymorphic]
```
```{r methods, eval=FALSE}
methods(class=class(phiX174Phage)) # 'DNAStringSet' methods
```
**Exercises**
1. Load the [Biostrings][] package and phiX174Phage data set. What class
is phiX174Phage? Find the help page for the class, and identify
interesting functions that apply to it.
2. Discover vignettes in the Biostrings package with
`vignette(package="Biostrings")`. Add another argument to the
`vignette` function to view the 'BiostringsQuickOverview' vignette.
3. If the internet is available, navigate to the Biostrings landing
page on http://bioconductor.org. Do this by visiting the
[biocViews][] page. Can you find the BiostringsQuickOverview
vignette on the web site?
4. The following code loads some sample data, 6 versions of the
phiX174Phage genome as a DNAStringSet object.
```{r phiX}
library(Biostrings)
data(phiX174Phage)
```
Explain what the following code does, and how it works
```{r consensusMatrix}
m <- consensusMatrix(phiX174Phage)[1:4,]
polymorphic <- which(colSums(m != 0) > 1)
mapply(substr, polymorphic, polymorphic, MoreArgs=list(x=phiX174Phage))
```
# Working with genomic ranges
## _IRanges_ and _GRanges_
The [IRanges][] package defines an important class for specifying
integer ranges, e.g.,
```{r iranges}
library(IRanges)
ir <- IRanges(start=c(10, 20, 30), width=5)
ir
```
There are many interesting operations to be performed on ranges, e.g,
`flank()` identifies adjacent ranges
```{r iranges-flank}
flank(ir, 3)
```
The `IRanges` class is part of a class hierarchy. To see this, ask R for
the class of `ir`, and for the class definition of the `IRanges` class
```{r iranges-class}
class(ir)
getClass(class(ir))
```
Notice that `IRanges` extends the `Ranges` class. Now try entering
`?flank` (`?"flank,"` if not using _RStudio_, where `` means
to press the tab key to ask for tab completion). You can see that
there are help pages for `flank` operating on several different
classes. Select the completion
```{r iranges-flank-method, eval=FALSE}
?"flank,Ranges-method"
```
and verify that you're at the page that describes the method relevant
to an `IRanges` instance. Explore other range-based operations.
The [GenomicRanges][] package extends the notion of ranges to include
features relevant to application of ranges in sequence analysis,
particularly the ability to associate a range with a sequence name
(e.g., chromosome) and a strand. Create a `GRanges` instance based on
our `IRanges` instance, as follows
```{r granges}
library(GenomicRanges)
gr <- GRanges(c("chr1", "chr1", "chr2"), ir, strand=c("+", "-", "+"))
gr
```
The notion of flanking sequence has a more nuanced meaning in
biology. In particular we might expect that flanking sequence on the
`+` strand would precede the range, but on the minus strand would
follow it. Verify that `flank` applied to a `GRanges` object has this
behavior.
```{r granges-flank}
flank(gr, 3)
```
Discover what classes `GRanges` extends, find the help page
documenting the behavior of `flank` when applied to a `GRanges` object,
and verify that the help page documents the behavior we just observed.
```{r granges-class}
class(gr)
getClass(class(gr))
```
```{r granges-flank-method, eval=FALSE}
?"flank,GenomicRanges-method"
```
Notice that the available `flank()` methods have been augmented by the
methods defined in the _GenomicRanges_ package.
It seems like there might be a number of helpful methods available for
working with genomic ranges; we can discover some of these from the
command line, indicating that the methods should be on the current
`search()` path
```{r granges-methods, eval=FALSE}
showMethods(class="GRanges", where=search())
```
Use `help()` to list the help pages in the `GenomicRanges` package,
and `vignettes()` to view and access available vignettes; these are
also available in the RStudio 'Help' tab.
```{r granges-man-and-vignettes, eval=FALSE}
help(package="GenomicRanges")
vignette(package="GenomicRanges")
vignette(package="GenomicRanges", "GenomicRangesHOWTOs")
```
## Range-based operations
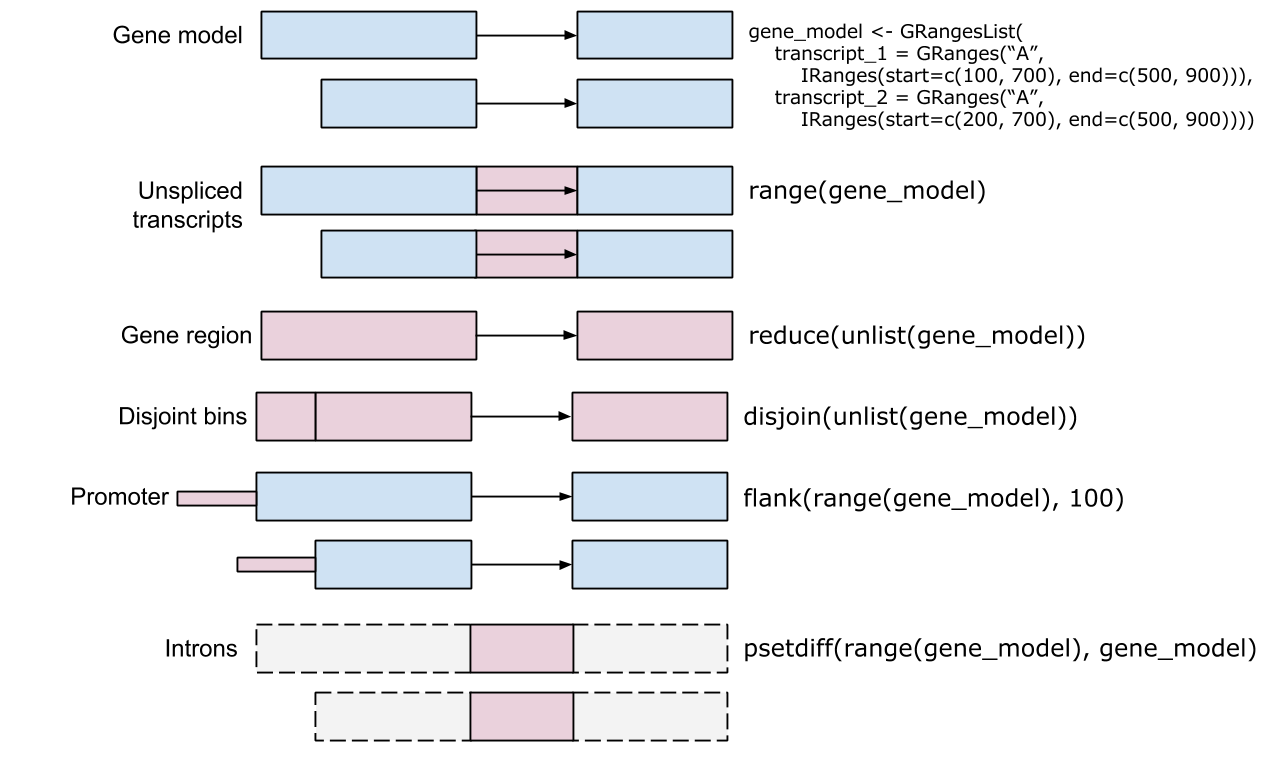
Ranges
- IRanges
- `start()` / `end()` / `width()`
- Vector-like -- `length()`, subset, etc.
- 'metadata', `mcols()`
- GRanges
- 'seqnames' (chromosome), 'strand'
- `Seqinfo`, including `seqlevels` and `seqlengths`
Intra-range methods
- Independent of other ranges in the same object
- GRanges variants strand-aware
- `shift()`, `narrow()`, `flank()`, `promoters()`, `resize()`,
`restrict()`, `trim()`
- See `?"intra-range-methods"`
Inter-range methods
- Depends on other ranges in the same object
- `range()`, `reduce()`, `gaps()`, `disjoin()`
- `coverage()` (!)
- see `?"inter-range-methods"`
Between-range methods
- Functions of two (or more) range objects
- `findOverlaps()`, `countOverlaps()`, ..., `%over%`, `%within%`,
`%outside%`; `union()`, `intersect()`, `setdiff()`, `punion()`,
`pintersect()`, `psetdiff()`
IRangesList, GRangesList
- List: all elements of the same type
- Many *List-aware methods, but a common 'trick': apply a vectorized
function to the unlisted representaion, then re-list
grl <- GRangesList(...)
orig_gr <- unlist(grl)
transformed_gr <- FUN(orig)
transformed_grl <- relist(transformed_gr, grl)
# How to use high-throughput sequence data types in _Bioconductor_
The following sections briefly summarize some of the most important
file types in high-throughput sequence analysis. _Briefly_ review
these, or those that are most relevant to your research, before
starting on the section [Data Representation in _R_ /
_Bioconductor_](#data-representation-in-r-bioconductor)
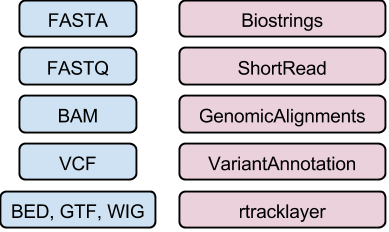
## DNA / amino acid sequences: FASTA files
Input & manipulation: [Biostrings][]
>NM_078863_up_2000_chr2L_16764737_f chr2L:16764737-16766736
gttggtggcccaccagtgccaaaatacacaagaagaagaaacagcatctt
gacactaaaatgcaaaaattgctttgcgtcaatgactcaaaacgaaaatg
...
atgggtatcaagttgccccgtataaaaggcaagtttaccggttgcacggt
>NM_001201794_up_2000_chr2L_8382455_f chr2L:8382455-8384454
ttatttatgtaggcgcccgttcccgcagccaaagcactcagaattccggg
cgtgtagcgcaacgaccatctacaaggcaatattttgatcgcttgttagg
...
## Reads: FASTQ files
Input & manipulation: [ShortRead][] `readFastq()`, `FastqStreamer()`,
`FastqSampler()`
@ERR127302.1703 HWI-EAS350_0441:1:1:1460:19184#0/1
CCTGAGTGAAGCTGATCTTGATCTACGAAGAGAGATAGATCTTGATCGTCGAGGAGATGCTGACCTTGACCT
+
HHGHHGHHHHHHHHDGG>CE?=896=:
@ERR127302.1704 HWI-EAS350_0441:1:1:1460:16861#0/1
GCGGTATGCTGGAAGGTGCTCGAATGGAGAGCGCCAGCGCCCCGGCGCTGAGCCGCAGCCTCAGGTCCGCCC
+
DE?DD>ED4>EEE>DE8EEEDE8B?EB<@3;BA79?,881B?@73;1?########################
- Quality scores: 'phred-like', encoded. See
[wikipedia](http://en.wikipedia.org/wiki/FASTQ_format#Encoding)
## Aligned reads: BAM files (e.g., ERR127306_chr14.bam)
Input & manipulation: 'low-level' [Rsamtools][], `scanBam()`,
`BamFile()`; 'high-level' [GenomicAlignments][]
- Header
@HD VN:1.0 SO:coordinate
@SQ SN:chr1 LN:249250621
@SQ SN:chr10 LN:135534747
@SQ SN:chr11 LN:135006516
...
@SQ SN:chrY LN:59373566
@PG ID:TopHat VN:2.0.8b CL:/home/hpages/tophat-2.0.8b.Linux_x86_64/tophat --mate-inner-dist 150 --solexa-quals --max-multihits 5 --no-discordant --no-mixed --coverage-search --microexon-search --library-type fr-unstranded --num-threads 2 --output-dir tophat2_out/ERR127306 /home/hpages/bowtie2-2.1.0/indexes/hg19 fastq/ERR127306_1.fastq fastq/ERR127306_2.fastq
- Alignments: ID, flag, alignment and mate
ERR127306.7941162 403 chr14 19653689 3 72M = 19652348 -1413 ...
ERR127306.22648137 145 chr14 19653692 1 72M = 19650044 -3720 ...
ERR127306.933914 339 chr14 19653707 1 66M120N6M = 19653686 -213 ...
ERR127306.11052450 83 chr14 19653707 3 66M120N6M = 19652348 -1551 ...
ERR127306.24611331 147 chr14 19653708 1 65M120N7M = 19653675 -225 ...
ERR127306.2698854 419 chr14 19653717 0 56M120N16M = 19653935 290 ...
ERR127306.2698854 163 chr14 19653717 0 56M120N16M = 19653935 2019 ...
- Alignments: sequence and quality
... GAATTGATCAGTCTCATCTGAGAGTAACTTTGTACCCATCACTGATTCCTTCTGAGACTGCCTCCACTTCCC *'%%%%%#&&%''#'&%%%)&&%%$%%'%%'&*****$))$)'')'%)))&)%%%%$'%%%%&"))'')%))
... TTGATCAGTCTCATCTGAGAGTAACTTTGTACCCATCACTGATTCCTTCTGAGACTGCCTCCACTTCCCCAG '**)****)*'*&*********('&)****&***(**')))())%)))&)))*')&***********)****
... TGAGAGTAACTTTGTACCCATCACTGATTCCTTCTGAGACTGCCTCCACTTCCCCAGCAGCCTCTGGTTTCT '******&%)&)))&")')'')'*((******&)&'')'))$))'')&))$)**&&****************
... TGAGAGTAACTTTGTACCCATCACTGATTCCTTCTGAGACTGCCTCCACTTCCCCAGCAGCCTCTGGTTTCT ##&&(#')$')'%&&#)%$#$%"%###&!%))'%%''%'))&))#)&%((%())))%)%)))%*********
... GAGAGTAACTTTGTACCCATCACTGATTCCTTCTGAGACTGCCTCCACTTCCCCAGCAGCCTCTGGTTTCTT )&$'$'$%!&&%&&#!'%'))%''&%'&))))''$""'%'%&%'#'%'"!'')#&)))))%$)%)&'"')))
... TTTGTACCCATCACTGATTCCTTCTGAGACTGCCTCCACTTCCCCAGCAGCCTCTGGTTTCTTCATGTGGCT ++++++++++++++++++++++++++++++++++++++*++++++**++++**+**''**+*+*'*)))*)#
... TTTGTACCCATCACTGATTCCTTCTGAGACTGCCTCCACTTCCCCAGCAGCCTCTGGTTTCTTCATGTGGCT ++++++++++++++++++++++++++++++++++++++*++++++**++++**+**''**+*+*'*)))*)#
- Alignments: Tags
... AS:i:0 XN:i:0 XM:i:0 XO:i:0 XG:i:0 NM:i:0 MD:Z:72 YT:Z:UU NH:i:2 CC:Z:chr22 CP:i:16189276 HI:i:0
... AS:i:0 XN:i:0 XM:i:0 XO:i:0 XG:i:0 NM:i:0 MD:Z:72 YT:Z:UU NH:i:3 CC:Z:= CP:i:19921600 HI:i:0
... AS:i:0 XN:i:0 XM:i:0 XO:i:0 XG:i:0 NM:i:4 MD:Z:72 YT:Z:UU XS:A:+ NH:i:3 CC:Z:= CP:i:19921465 HI:i:0
... AS:i:0 XN:i:0 XM:i:0 XO:i:0 XG:i:0 NM:i:4 MD:Z:72 YT:Z:UU XS:A:+ NH:i:2 CC:Z:chr22 CP:i:16189138 HI:i:0
... AS:i:0 XN:i:0 XM:i:0 XO:i:0 XG:i:0 NM:i:5 MD:Z:72 YT:Z:UU XS:A:+ NH:i:3 CC:Z:= CP:i:19921464 HI:i:0
... AS:i:0 XM:i:0 XO:i:0 XG:i:0 MD:Z:72 NM:i:0 XS:A:+ NH:i:5 CC:Z:= CP:i:19653717 HI:i:0
... AS:i:0 XM:i:0 XO:i:0 XG:i:0 MD:Z:72 NM:i:0 XS:A:+ NH:i:5 CC:Z:= CP:i:19921455 HI:i:1
## Called variants: VCF files
Input and manipulation: [VariantAnnotation][] `readVcf()`,
`readInfo()`, `readGeno()` selectively with `ScanVcfParam()`.
- Header
##fileformat=VCFv4.2
##fileDate=20090805
##source=myImputationProgramV3.1
##reference=file:///seq/references/1000GenomesPilot-NCBI36.fasta
##contig=
##phasing=partial
##INFO=
##INFO=
...
##FILTER=
##FILTER=
...
##FORMAT=
##FORMAT=
- Location
#CHROM POS ID REF ALT QUAL FILTER ...
20 14370 rs6054257 G A 29 PASS ...
20 17330 . T A 3 q10 ...
20 1110696 rs6040355 A G,T 67 PASS ...
20 1230237 . T . 47 PASS ...
20 1234567 microsat1 GTC G,GTCT 50 PASS ...
- Variant INFO
#CHROM POS ... INFO ...
20 14370 ... NS=3;DP=14;AF=0.5;DB;H2 ...
20 17330 ... NS=3;DP=11;AF=0.017 ...
20 1110696 ... NS=2;DP=10;AF=0.333,0.667;AA=T;DB ...
20 1230237 ... NS=3;DP=13;AA=T ...
20 1234567 ... NS=3;DP=9;AA=G ...
- Genotype FORMAT and samples
... POS ... FORMAT NA00001 NA00002 NA00003
... 14370 ... GT:GQ:DP:HQ 0|0:48:1:51,51 1|0:48:8:51,51 1/1:43:5:.,.
... 17330 ... GT:GQ:DP:HQ 0|0:49:3:58,50 0|1:3:5:65,3 0/0:41:3
... 1110696 ... GT:GQ:DP:HQ 1|2:21:6:23,27 2|1:2:0:18,2 2/2:35:4
... 1230237 ... GT:GQ:DP:HQ 0|0:54:7:56,60 0|0:48:4:51,51 0/0:61:2
... 1234567 ... GT:GQ:DP 0/1:35:4 0/2:17:2 1/1:40:3
## Genome annotations: BED, WIG, GTF, etc. files
Input: [rtracklayer][] `import()`
- BED: range-based annotation (see
http://genome.ucsc.edu/FAQ/FAQformat.html for definition of this and
related formats)
- WIG / bigWig: dense, continuous-valued data
- GTF: gene model
- Component coordinates
7 protein_coding gene 27221129 27224842 . - . ...
...
7 protein_coding transcript 27221134 27224835 . - . ...
7 protein_coding exon 27224055 27224835 . - . ...
7 protein_coding CDS 27224055 27224763 . - 0 ...
7 protein_coding start_codon 27224761 27224763 . - 0 ...
7 protein_coding exon 27221134 27222647 . - . ...
7 protein_coding CDS 27222418 27222647 . - 2 ...
7 protein_coding stop_codon 27222415 27222417 . - 0 ...
7 protein_coding UTR 27224764 27224835 . - . ...
7 protein_coding UTR 27221134 27222414 . - . ...
- Annotations
gene_id "ENSG00000005073"; gene_name "HOXA11"; gene_source "ensembl_havana"; gene_biotype "protein_coding";
...
... transcript_id "ENST00000006015"; transcript_name "HOXA11-001"; transcript_source "ensembl_havana"; tag "CCDS"; ccds_id "CCDS5411";
... exon_number "1"; exon_id "ENSE00001147062";
... exon_number "1"; protein_id "ENSP00000006015";
... exon_number "1";
... exon_number "2"; exon_id "ENSE00002099557";
... exon_number "2"; protein_id "ENSP00000006015";
... exon_number "2";
...
...
# Exercises: Data representation in _R_ / _Bioconductor_
This section briefly illustrates how different high-throughput
sequence data types are represented in _R_ / _Bioconductor_. Select
relevant data types for your area of interest, and work through the
examples. Take time to consult help pages, understand the output of
function calls, and the relationship between standard data formats
(summarized in the previous section) and the corresponding _R_ /
_Bioconductor_ representation.
## _Biostrings_ (DNA or amino acid sequences)
Classes
- XString, XStringSet, e.g., DNAString (genomes),
DNAStringSet (reads)
Methods --
- [Cheat sheat](http://bioconductor.org/packages/release/bioc/vignettes/Biostrings/inst/doc/BiostringsQuickOverview.pdf)
- Manipulation, e.g., `reverseComplement()`
- Summary, e.g., `letterFrequency()`
- Matching, e.g., `matchPDict()`, `matchPWM()`
Related packages
- [BSgenome][]
- Whole-genome representations
- Model and custom
- [ShortRead][]
- FASTQ files
Example
- Whole-genome sequences are distrubuted by ENSEMBL, NCBI, and others
as FASTA files; model organism whole genome sequences are packaged
into more user-friendly `BSgenome` packages. The following
calculates GC content across chr14.
```{r BSgenome-require, message=FALSE}
library(BSgenome.Hsapiens.UCSC.hg38)
chr14_range = GRanges("chr14", IRanges(1, seqlengths(Hsapiens)["chr14"]))
chr14_dna <- getSeq(Hsapiens, chr14_range)
letterFrequency(chr14_dna, "GC", as.prob=TRUE)
```
**Exercises**
0. Setup
- Mouse CDS sequence, from Ensembl: https://useast.ensembl.org/info/data/ftp/index.html
```{r}
library(Biostrings)
url <- "ftp://ftp.ensembl.org/pub/release-92/fasta/mus_musculus/cds/Mus_musculus.GRCm38.cds.all.fa.gz"
fl <- BiocFileCache::bfcrpath(rnames = url)
cds <- rtracklayer::import(fl, "fasta")
```
1. For simplicity, clean up the data to remove cds with width not a
multiple of three. Remove cds that don't start with a start codon
`ATG` or end with a stop codon `c("TAA", "TAG", "TGA")`
```{r}
pred1 <- width(cds) %% 3 == 0
table(pred1)
pred2 <- narrow(cds, 1, 3) == "ATG"
stops <- c("TAA", "TAG", "TGA")
pred3 <- narrow(cds, width(cds) - 2, width(cds)) %in% stops
table(pred1 & pred2 & pred3)
cds <- cds[ pred1 & pred2 & pred3 ]
```
2. What does the distribution of widths of the cds look like? Which
cds has maximum width?
```{r}
hist(log10(width(cds)))
cds[ which.max(width(cds)) ]
names(cds)[ which.max(width(cds)) ]
```
3. Use `letterFrequency()` to calculate the GC content of each cds;
visualize the distribution of GC content.
```{r}
gc <- letterFrequency(cds, "GC", as.prob=TRUE)
head(gc)
hist(gc)
plot( log10(width(cds)), gc, pch=".")
```
4. Summarize codon usage in each CDS. Which codons are used most
frequently over all CDS?
```{r}
AMINO_ACID_CODE
aa <- translate(cds)
codon_use <- letterFrequency(aa, names(AMINO_ACID_CODE))
head(codon_use)
```
5. (Advanced) -- `DNAStringSet` inherits from `Vector` and
`Annotated`, which means that each element (sequence) can have
additional information, for instance we can associate GC content
with each sequence
```{r}
mcols(cds) <- DataFrame(
GC = gc[,"G|C"]
)
mcols(cds, use.names = FALSE)
mcols(cds[1:3], use.names = FALSE)
```
## _SummarizedExperiment_
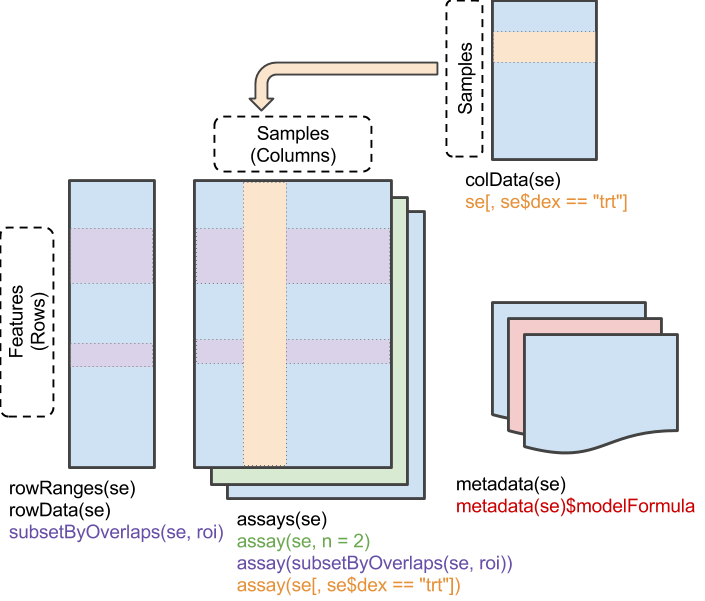
Motivation: reproducible & interoperable
- Matrix of feature x sample measurements, `assays()`
- Addition description about samples, `colData()`
- Covariates, e.g., age, gender
- Experimental design, e.g., treatment group
- Additional information about features, `rowData()`
- Gene names, width, GC content, ...
- Genomic ranges(!)
- Derived values, E.g., log-fold change between treatments, _P_-value, ...
- Information about the experiment as a whole -- `metadata()`
Example 1: Bulk RNA-seq `airway` data
- Attach the airway library and data set
```{r}
library(airway)
data(airway)
airway
```
- Explore the phenotypic data describing samples. Subset to include just the `"untrt"` samples.
```{r}
colData(airway)
airway[ , airway$dex == "untrt"]
```
- Calculate library size as the column sums of the assays. Reflect on
the relationship between library size and cell / dex column
variables and consequences for differential expression analysis.
```{r}
colSums(assay(airway))
```
Example 2 (advanced): single-cell RNA-seq.
- Hemberg lab [scRNA-seq Datasets][] 'Manno' [Mouse brain data set (rds)][]
```{r, message=FALSE}
url <- "https://scrnaseq-public-datasets.s3.amazonaws.com/scater-objects/manno_mouse.rds"
fl <- BiocFileCache::bfcrpath(rnames = url)
sce <- readRDS(fl)
```
**Exercises**
- What's the frequency of 0 counts in the single cell assay data?
- What's the distribution of library sizes in the single cell assay data?
- Create a random sample of 100 cells and visualize the relationship
between samples using `dist()` and `cmdscale()`.
- can you identify what column of `colData()` is responsible for any
pattern you see?
- In exploring the covariates, are the possible problems with confounding?
[scRNA-seq Datasets]: https://hemberg-lab.github.io/scRNA.seq.datasets/
[Mouse brain data set (rds)]: https://scrnaseq-public-datasets.s3.amazonaws.com/scater-objects/manno_mouse.rds
## _GenomicRanges_
Example
```{r ranges, message=FALSE}
library(GenomicRanges)
gr <- GRanges(c("chr1:10-14:+", "chr1:20-24:+", "chr1:22-26:+"))
shift(gr, 1) # 1-based coordinates!
range(gr) # intra-range
reduce(gr) # inter-range
coverage(gr)
setdiff(range(gr), gr) # 'introns'
```
**Exercises**
1. Which of my SNPs overlap genes?
```{r}
genes <- GRanges(c("chr1:30-40:+", "chr1:60-70:-"))
snps <- GRanges(c("chr1:35", "chr1:60", "chr1:45"))
countOverlaps(snps, genes) > 0
```
2. Which gene is 'nearest' my regulatory region? Which gene does my
regulatory region _precede_ (i.e., upstream of)
```{r}
reg <- GRanges(c("chr1:50-55", "chr1:75-80"))
nearest(reg, genes)
precede(reg, genes)
```
3. What range do short reads cover? depth of coverage?
```{r}
reads <- GRanges(c("chr1:10-19", "chr1:15-24", "chr1:30-41"))
coverage(reads, width = 100)
as(coverage(reads, width = 100), "GRanges")
```
Reference
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, et al. (2013)
Software for Computing and Annotating Genomic Ranges. PLoS Comput
Biol 9(8): e1003118. doi:10.1371/journal.pcbi.1003118
## _GenomicAlignments_ (Aligned reads)
Classes -- GenomicRanges-like behaivor
- GAlignments, GAlignmentPairs, GAlignmentsList
Methods
- `readGAlignments()`, `readGAlignmentsList()`
- Easy to restrict input, iterate in chunks
- `summarizeOverlaps()`
**Exercises**
1. Find reads supporting the junction identified above, at position
19653707 + 66M = 19653773 of chromosome 14
```{r bam-require}
library(GenomicRanges)
library(GenomicAlignments)
library(Rsamtools)
## our 'region of interest'
roi <- GRanges("chr14", IRanges(19653773, width=1))
## sample data
library('RNAseqData.HNRNPC.bam.chr14')
bf <- BamFile(RNAseqData.HNRNPC.bam.chr14_BAMFILES[[1]], asMates=TRUE)
## alignments, junctions, overlapping our roi
paln <- readGAlignmentsList(bf)
j <- summarizeJunctions(paln, with.revmap=TRUE)
j_overlap <- j[j %over% roi]
## supporting reads
paln[j_overlap$revmap[[1]]]
```
## _VariantAnnotation_ (Called variants)
Classes -- GenomicRanges-like behavior
- VCF -- 'wide'
- VRanges -- 'tall'
Functions and methods
- I/O and filtering: `readVcf()`, `readGeno()`, `readInfo()`,
`readGT()`, `writeVcf()`, `filterVcf()`
- Annotation: `locateVariants()` (variants overlapping ranges),
`predictCoding()`, `summarizeVariants()`
- SNPs: `genotypeToSnpMatrix()`, `snpSummary()`
**Exerises**
1. Read variants from a VCF file, and annotate with respect to a known
gene model
```{r vcf, message=FALSE}
## input variants
library(VariantAnnotation)
fl <- system.file("extdata", "chr22.vcf.gz", package="VariantAnnotation")
vcf <- readVcf(fl, "hg19")
seqlevels(vcf) <- "chr22"
## known gene model
library(TxDb.Hsapiens.UCSC.hg19.knownGene)
coding <- locateVariants(rowRanges(vcf),
TxDb.Hsapiens.UCSC.hg19.knownGene,
CodingVariants())
head(coding)
```
Related packages
- [ensemblVEP][]
- Forward variants to Ensembl Variant Effect Predictor
- [VariantTools][], [h5vc][]
- Call variants
Reference
- Obenchain, V, Lawrence, M, Carey, V, Gogarten, S, Shannon, P, and
Morgan, M. VariantAnnotation: a Bioconductor package for exploration
and annotation of genetic variants. Bioinformatics, first published
online March 28, 2014
[doi:10.1093/bioinformatics/btu168](http://bioinformatics.oxfordjournals.org/content/early/2014/04/21/bioinformatics.btu168)
## _rtracklayer_ (Genome annotations)
- Import BED, GTF, WIG, etc
- Export GRanges to BED, GTF, WIG, ...
- Access UCSC genome browser
# Extended exercises
## Summarize overlaps
The goal is to count the number of reads overlapping exons grouped
into genes. This type of count data is the basic input for RNASeq
differential expression analysis, e.g., through [DESeq2][] and
[edgeR][].
1. Identify the regions of interest. We use a 'TxDb' package with gene
models already defined; the genome (hg19) is determined by the
genome used for read alignment in the sample BAM files.
```{r summarizeOverlaps-roi, message=FALSE}
library(TxDb.Hsapiens.UCSC.hg19.knownGene)
exByGn <- exonsBy(TxDb.Hsapiens.UCSC.hg19.knownGene, "gene")
## only chromosome 14
seqlevels(exByGn, pruning.mode="coarse") = "chr14"
```
2. Identify the sample BAM files.
```{r summarizeOverlaps-bam, message=FALSE}
library(RNAseqData.HNRNPC.bam.chr14)
length(RNAseqData.HNRNPC.bam.chr14_BAMFILES)
```
3. Summarize overlaps, optionally in parallel
```{r summarizeOverlaps}
## next 2 lines optional; non-Windows
library(BiocParallel)
register(MulticoreParam(workers=detectCores()))
olaps <- summarizeOverlaps(exByGn, RNAseqData.HNRNPC.bam.chr14_BAMFILES)
```
4. Explore our handiwork, e.g., library sizes (column sums),
relationship between gene length and number of mapped reads, etc.
```{r summarizeOverlaps-explore}
olaps
head(assay(olaps))
colSums(assay(olaps)) # library sizes
plot(sum(width(olaps)), rowMeans(assay(olaps)), log="xy")
```
5. As an advanced exercise, investigate the relationship between GC
content and read count
```{r summarizeOverlaps-gc}
library(BSgenome.Hsapiens.UCSC.hg19)
sequences <- getSeq(BSgenome.Hsapiens.UCSC.hg19, rowRanges(olaps))
gcPerExon <- letterFrequency(unlist(sequences), "GC")
gc <- relist(as.vector(gcPerExon), sequences)
gc_percent <- sum(gc) / sum(width(olaps))
plot(gc_percent, rowMeans(assay(olaps)), log="y")
```
[biocViews]: http://bioconductor.org/packages/release/BiocViews.html#___Software
[AnnotationData]: http://bioconductor.org/packages/release/BiocViews.html#___AnnotationData
[aprof]: http://cran.r-project.org/web/packages/aprof/index.html
[hexbin]: http://cran.r-project.org/web/packages/hexbin/index.html
[lineprof]: https://github.com/hadley/lineprof
[microbenchmark]: http://cran.r-project.org/web/packages/microbenchmark/index.html
[AnnotationDbi]: http://bioconductor.org/packages/AnnotationDbi
[BSgenome]: http://bioconductor.org/packages/BSgenome
[BiocParallel]: http://bioconductor.org/packages/BiocParallel
[Biostrings]: http://bioconductor.org/packages/Biostrings
[CNTools]: http://bioconductor.org/packages/CNTools
[ChIPQC]: http://bioconductor.org/packages/ChIPQC
[ChIPpeakAnno]: http://bioconductor.org/packages/ChIPpeakAnno
[DESeq2]: http://bioconductor.org/packages/DESeq2
[DiffBind]: http://bioconductor.org/packages/DiffBind
[GenomicAlignments]: http://bioconductor.org/packages/GenomicAlignments
[GenomicRanges]: http://bioconductor.org/packages/GenomicRanges
[IRanges]: http://bioconductor.org/packages/IRanges
[KEGGREST]: http://bioconductor.org/packages/KEGGREST
[PSICQUIC]: http://bioconductor.org/packages/PSICQUIC
[rtracklayer]: http://bioconductor.org/packages/rtracklayer
[Rsamtools]: http://bioconductor.org/packages/Rsamtools
[ShortRead]: http://bioconductor.org/packages/ShortRead
[VariantAnnotation]: http://bioconductor.org/packages/VariantAnnotation
[VariantFiltering]: http://bioconductor.org/packages/VariantFiltering
[VariantTools]: http://bioconductor.org/packages/VariantTools
[biomaRt]: http://bioconductor.org/packages/biomaRt
[cn.mops]: http://bioconductor.org/packages/cn.mops
[h5vc]: http://bioconductor.org/packages/h5vc
[edgeR]: http://bioconductor.org/packages/edgeR
[ensemblVEP]: http://bioconductor.org/packages/ensemblVEP
[limma]: http://bioconductor.org/packages/limma
[metagenomeSeq]: http://bioconductor.org/packages/metagenomeSeq
[phyloseq]: http://bioconductor.org/packages/phyloseq
[snpStats]: http://bioconductor.org/packages/snpStats
[org.Hs.eg.db]: http://bioconductor.org/packages/org.Hs.eg.db
[TxDb.Hsapiens.UCSC.hg38.knownGene]: http://bioconductor.org/packages/TxDb.Hsapiens.UCSC.hg38.knownGene
[BSgenome.Hsapiens.UCSC.hg38]: http://bioconductor.org/packages/BSgenome.Hsapiens.UCSC.hg38
# End matter
## Session Info
```{r}
sessionInfo()
```
## Acknowledgements
Research reported in this tutorial was supported by the National Human
Genome Research Institute and the National Cancer Institute of the
National Institutes of Health under award numbers U41HG004059 and
U24CA180996.
This project has received funding from the European Research Council
(ERC) under the European Union's Horizon 2020 research and innovation
programme (grant agreement number 633974)